Google Gen AI Course Project - Medical Assistant v1
Designed and Implemented a Medical Assistant that can provide medical Q&As and doctor search.
Project Description
In this project, a Medical Assistant chatbot is implemented that can answer general medical Q&As and search for doctors based on the user queries. This enables smooth communication with the user, while allowing accessing domain knowledge of the medical field through RAG system with vector database storing medical documents.
This project utilizes the GenAI agent method provided by Gemini API to allow grounding the information by Google Search, and accessing online doctor database for making medical appointment.
The project can be found at: https://www.kaggle.com/code/yinglongmiao/medical-assistant-v1. To try it out, go to the “Running Example” section and interacts with the chatbot through an input dialog!
Implementation
The flowchart of the system can be described as follows.
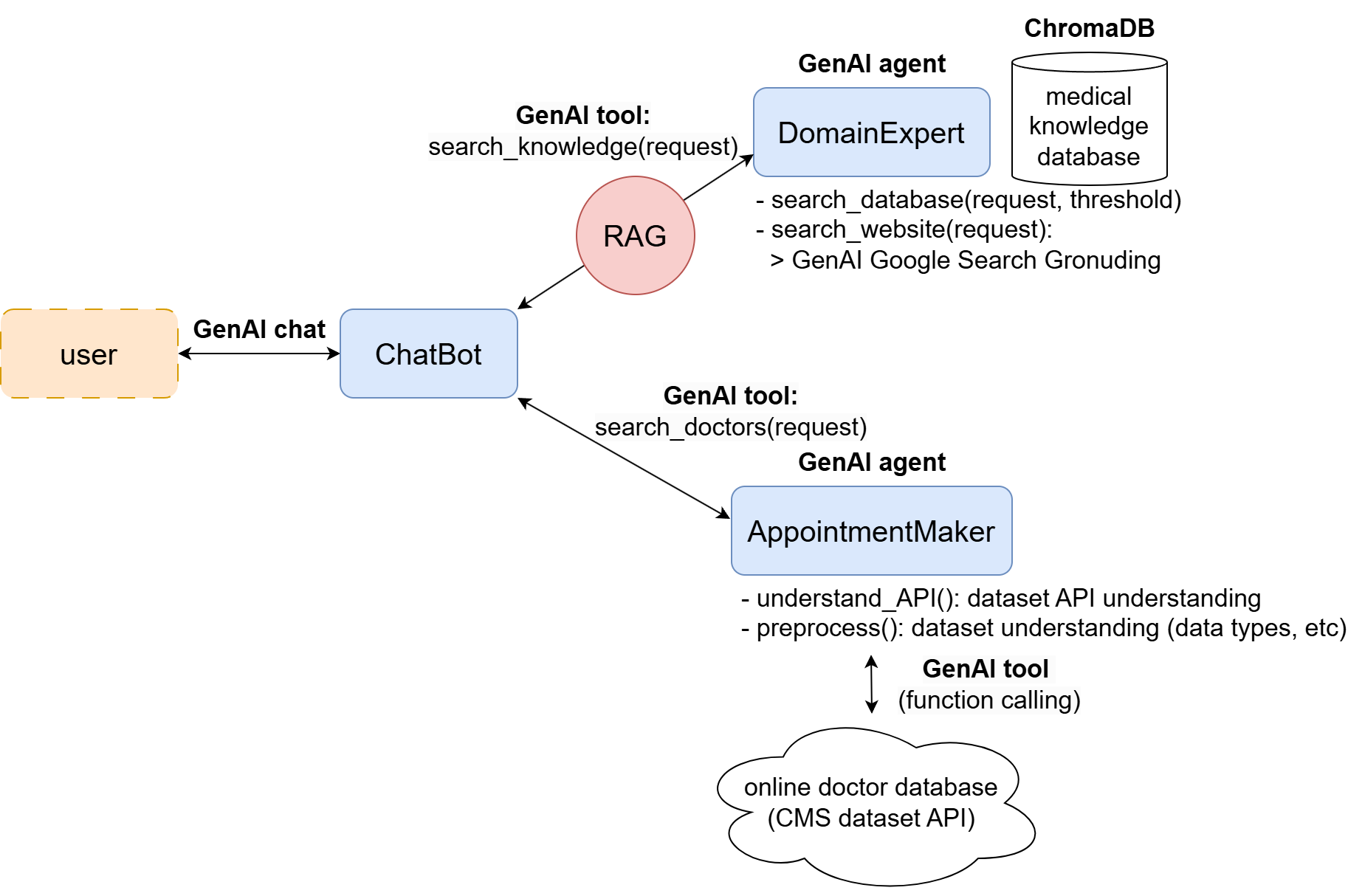
ChatBot
the ChatBot provides communication with the user through a chat session by Gemini API. The chatbot uses tools implemented by the domain expert and the appointment maker, as shown below:
self.tools = self.domain_expert.tools + self.appointment_maker.tools + [self.closing_app]
It includes an additional function self.closing_app which sets the status of the chatbot in case the user wants to terminate the session. This is controlled by a status variable self.TERMINATE.
The chat session is initialized using “gemini-2.0-flash” model in the constructor of the class.
self.chat_session = self.client.chats.create(
model="gemini-2.0-flash",
config=types.GenerateContentConfig(
system_instruction=self.system_instr,
tools=self.tools,
),
)
THe main function chat() implements the main flow of communication with the user.
def chat(self):
# * perform the basic I/O operation: getting inputs from the user, and perform actions accordingly
self.TERMINATE = False
while True:
user_input = input("User: ")
# If it looks like the user is trying to quit, flag the conversation
# as over.
# ref: https://www.kaggle.com/code/markishere/day-3-building-an-agent-with-langgraph
if user_input in {"q", "quit", "exit", "goodbye"}:
break
response = self.chat_session.send_message(user_input)
print(f"\n{response.text}")
if self.TERMINATE:
break
The user can input “q” and other ways to exit the chat session. At the same time, if the user input indicates he/she wants to quit the session, it is captured by the closing_app() function.
Domain Expert
The DomainExpert implements the functionalities to support accessing domain knowledge of the medical field through RAG (vector database) and Google Search.
It uses a ChromaDB to construct a vector database for RAG operation of extracting medical knowledge database. Cosine distance is used to facilitate similarity search at high dimensions.
embed_fn = self.GeminiEmbeddingFunction()
embed_fn.document_mode = True
chroma_client = chromadb.Client()
db = chroma_client.get_or_create_collection(name=DB_NAME, embedding_function=embed_fn, metadata={"hnsw:space": "cosine"}) # or "dotproduct")
The vector database uses the embedding function provided by the Gemini API, as shown below.
response = self.client.models.embed_content(
model="models/text-embedding-004",
contents=input,
config=types.EmbedContentConfig(
task_type=embedding_task,
),
)
When the tool search_knowledge(request) is executed, it first sees if information can be found in the database with high similarity (filtered by distance_threshold):
def search_database(self, request: str, distance_threshold: float = 0.8) -> list[str]:
self.embed_fn.document_mode = False # retrieval
result = self.db.query(query_texts=[request], n_results=1)
documents = result['documents'][0]
# * based on the similarity, determine if to search online or not.
documents = [documents[i] for i in range(len(documents)) if result['distances'][0][i] <= distance_threshold]
return documents
If no related information can be found (determined by the similarity), then the agent applies Google search. This is implemented by calling a GenAI call using Google search as grounding:
response = self.genai_client.models.generate_content(
model="gemini-2.0-flash",
contents=f"Please make a Google search for the user query: {request}",
config=types.GenerateContentConfig(
tools=[self.google_search_tool],
response_modalities=["TEXT"],
)
)
This is used as this has higher rate limit than calling Google Search function directly, and it enables smarter searches implemented by the API.
The hope is that I can add the information extracted from the website (stored in the response.candidates[0].grounding_metadata.grounding_chunks field) to the vector database. But due to time constraint, this is left for future work.
The agent then returns a response based on the documents, or Google search directly, to the ChatBot agent. Extracted documents from the database is added as response as well.
AppointmentMaker
The AppointmentMaker is responsible for searching for doctors and (future work) making appointments if needed. This is achieved by accessing information from the online database CMS National Downloadable File. Although the idea is easy, the implementation is a bit tricky due to several challenges:
- the API for accessing the online database is complex, and I’m not familiar with it. Hence an idea came out: what about using the GenAI to call the API directly? However, this leads to the next challenges.
- It is found out that Gemini API code execution does not allow Internet connection for security reasons. Hence I can only work locally. There are two ways to solve it: firstly I can download the dataset from online. But it is found that the dataset is too large and takes to much time to download. Another solution is writing functions to access the online API, while using the Gemini API to generate GET or POST requests. The code for calling remote API endpoint is shown below:
def search_cms_database_json(query: dict) -> dict: """ given query formated in JSON, query the online CMS dataset. Args: query: JSON format dictionary formatting the query according to the CMS dataset API. Returns: retrieved data from online CMS dataset in JSON format if success. Return None if query failed. """ query_url = f'https://data.cms.gov/provider-data/api/1/datastore/query/{distribution_id}' headers = {'Content-Type': 'application/json'} response = requests.post(query_url, json=query, headers=headers) data = None if response.status_code == 200: print('Query succeed.') data = response.json() else: print(f"Query failed: {response.status_code}") return dataAfter the distribution id is found, this can call the remote API for database queries.
- When using the latter option, I need to know the syntax of the GET or POST requests. Luckily the API provides online specification of the API at API Specification (if you’re interested, feel free to check it out.). However, the specification string is too long, and naively I met the issue of hitting the token length limit.
The last step is closer to the solution. To make sure we can use Gemini API function call to access the database while referring to the online specification, one solution is to use the Gemini client to summarize the related API signatures, and then use this to guide the further API calls. To accelerate the search process and improve user experience, we can accelerate the user query search by preprocessing, such as understanding the available data fields in the database. This requires a preprocessing step that interacts with the database first to extract useful information that can help acceleration future queries. Hence the solution includes two important components: API understanding, and preprocessing.
The API understanding code snippet can be found below:
def understand_API(self):
"""
given the api spec, extract useful information for specific API endpoint.
"""
prompt = f"""extract relavent information for querying the database using POST via '/provider-data/api/1/datastore/query/distributionID' endpoint from the API specification, where distributionID is already provided as {self.distribution_id}.
Provide specification of query input.
Provide one example of full query input for future aid.
API SPECIFICATOIN: \n{self.api_spec}"""
print("parsing online database API...")
response = genai_client.models.generate_content(
model='gemini-2.0-flash',
contents=prompt,
# config=types.GenerateContentConfig()
)
return response.text
This returns the API specification that we are interested in using. The preprocessing step can be found below:
def preprocess(self):
"""
preprocess the database. Obtain useful information such as the distribution_id, and the fields available for future use.
"""
prompt = f"""You are going to use the database at CMS to search for doctors of specific specialties and for making appointment.
You need to do the following to preprocess information that can be used for future usage.
- understand the database schema and its available data fields.
given the API specification, use the defined function tool 'search_cms_database_json' with query argument to extract useful information that could be used for future reference.
This function tool is using '/provider-data/api/1/datastore/query/distributionID' endpoint from the API specification, where distributionID is already provided as {self.distribution_id}.
You just need to specify the JSON query as input to the function.
provide the database information in a structured way.
API SPECIFICATION: \n{self.api_info}
Note:
- set "limit" field as otherwise it may extract all the data in the database.
"""
print("preprocessing online database...")
response = self.genai_call(prompt)
return response
After naive implementation, I met the issue that the syntax during API calling is roughly correct but could sometimes lead to errors due to minor syntax issues. To mitigate the issue, I found that prompt engineering with one-shot or few-shot learning helps greatly, by asking the Gemini agent to provide one example during API understanding. After this, the agent can successfully call online API calls.
Afterward, the agent works, but struggles to find doctors that are of certain specialties (such as words described by “heart” or “kidney”, which are not available in the database.) To mitigate the problem, prompt engineering helps again, by instructing the database to first check out the keywords provided in the database for specialties. The complication is that the API does not provide calls for getting unique database items. However, I found that after implementing the prompt engineering, the agent is able to successfully solve most search queries.
Below shows the latest version of the search_doctors(query) function, notice how I added some “NOTE” to instruct the agent to first list out the values that are in the database. This greatly improves the robustness of the system.
def search_doctors(self, query: str):
"""
search from CMS dataset: https://data.cms.gov/provider-data/dataset/mj5m-pzi6#api
given the user query (symptom, etc), search for doctors for the treatment.
Achieve this by Python code generation and calling, given the API description found at: https://data.cms.gov/provider-data/api/1/metastore/schemas/dataset/items/mj5m-pzi6/docs
"""
prompt = f"""The user wants to search for doctors given the query.
You are going to use information of the API specification to extract information of the doctor.
Use the defined function tool 'search_cms_database_json' with JSON query argument to make queries.
API SPECIFICATION: \n{api_info}\n
prior knowledge of the database: \n{preprocessed_info}\n
user query: \n{query}
NOTE:
- you may want to first list out set of values that are included in the database before searching for keyword. Make sure the keyword exists in the database. Setting "limit" to be 0 won't work in the API.
- if the keyword does not exist in the database, try searching what unique keywords are included in the database to find similar ones. You can do this at the beginning to aid later steps.
- Make sure to include the contact information of the doctors (their location, phone number, etc) in the response.
- Prioritize shorter queries first.
Return the searched information at the end.
Response:
"""
response = self.genai_call(prompt)
return response
Future Work
Due to time constraints, I am not able to implement all the components that were originally planned. This includes:
- adding website references to the vector database (Note: copywrite issue might occur)
- adding evaluation mechanism to qualitatively and quantitatively evaluates the agent system.
I found the second step essential, as although the agent seems to perform smoothly, there is much stochasticity in its output, which is hard to define and evaluate quantitatively. It is important to obtain metrics for evaluating the performance of the system, such as the success rate, and how much the system is “professional” enough to provide medical assistant. It is important to analyze how much the agent is truthful and can provide professional medical feedbacks, which is an essential part of building medical assistant.
Reference
- https://www.kaggle.com/code/markishere/day-2-document-q-a-with-rag
- https://www.kaggle.com/code/markishere/day-2-embeddings-and-similarity-scores
- https://www.kaggle.com/code/markishere/day-3-function-calling-with-the-gemini-api
- https://data.cms.gov/provider-data/dataset/mj5m-pzi6#api